Platinum Genomes
This tutorial highlights some of the analysis features available in the Mosaic platform. We will perform a deep dive into the Platinum Exomes Project.
Select the project
Once you've logged into Mosaic, you will see the public projects that you have access to. To start this tutorial, click on the Platinum Exomes project in the table. You will then be taken to the project's dashboard that summarizes the project.
Applications
Every project links to a host of genomic data files, typically BAM/CRAM and/or VCF files. One role that Mosaic plays is to act as a launchpad for applications that can analyze these files. If you click on Applications in the left menu will bring up a list of the available apps that can be launched. This list will change depending on which page you are on in Mosaic. For example, when you click on Applications from the project dashboard, you can choose apps that operate on project level data.

The only app available in the free version of Mosaic, that operates on project level data is multibam.iobio - a quality control tool that compares sequencing quality of all selected samples in a project. Since the Platinum Exomes project only has 4 samples, we will not focus on that app here. Multibam.iobio is more useful for projects that contain lots of samples.
There are more apps available to operate on sample, or pedigree level data. These apps can be accessed in one of two ways. First, click on Samples in the left menu and you will be presented with a table of the four samples in this project. At the end of each row in the table, you can click the launch icon, and the menu of apps will again open, but now there is a different set of apps available. Selecting any of the apps will launch the app for the selected sample.
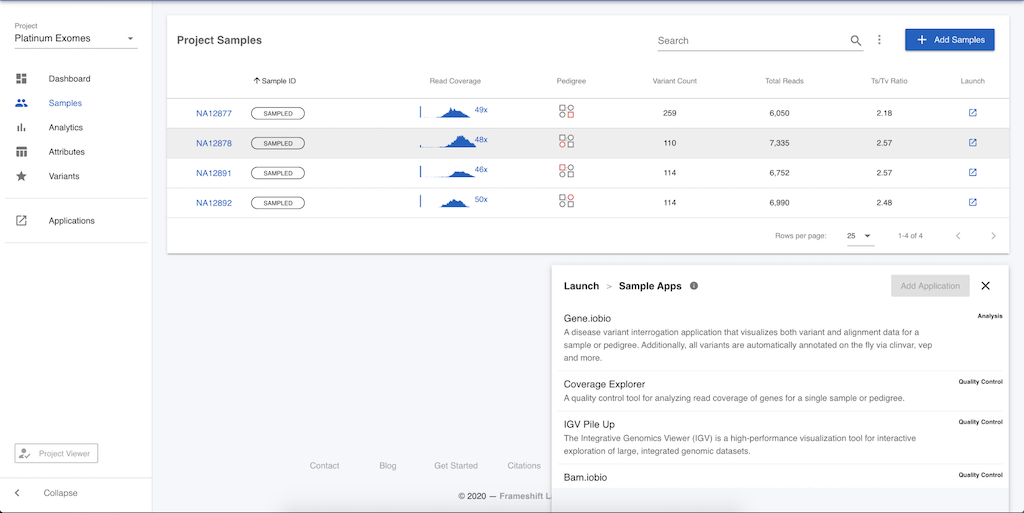
Alternatively, you can click on the sample name (e.g. NA12878) in the table and be taken to a page that summarizes information on this particular sample. Clicking on Applications in the left menu will again provide this list of apps available for analysing sample level data.
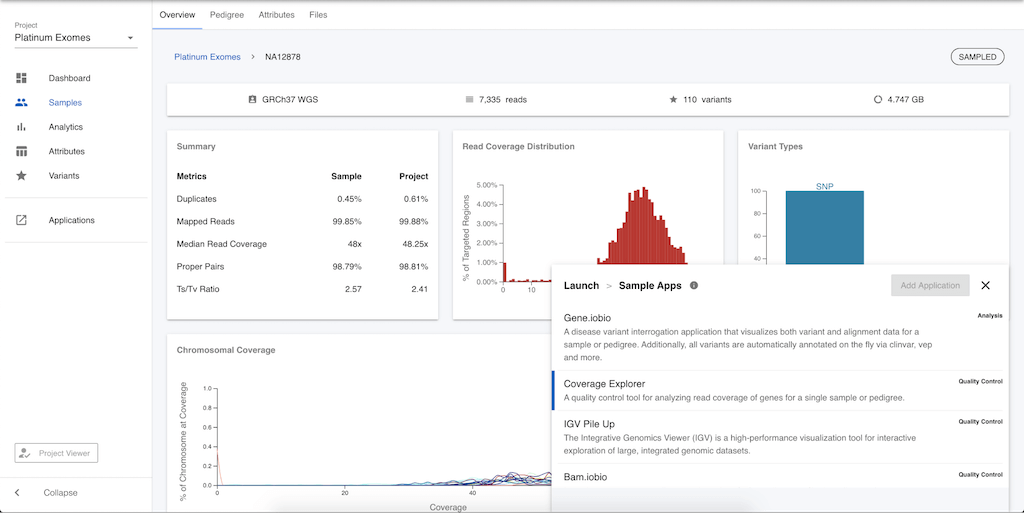
To demonstrate how these apps work, let's quickly take a look at the gene.iobio app. In the list of available apps, select gene.iobio, and you will be prompted to add more information.
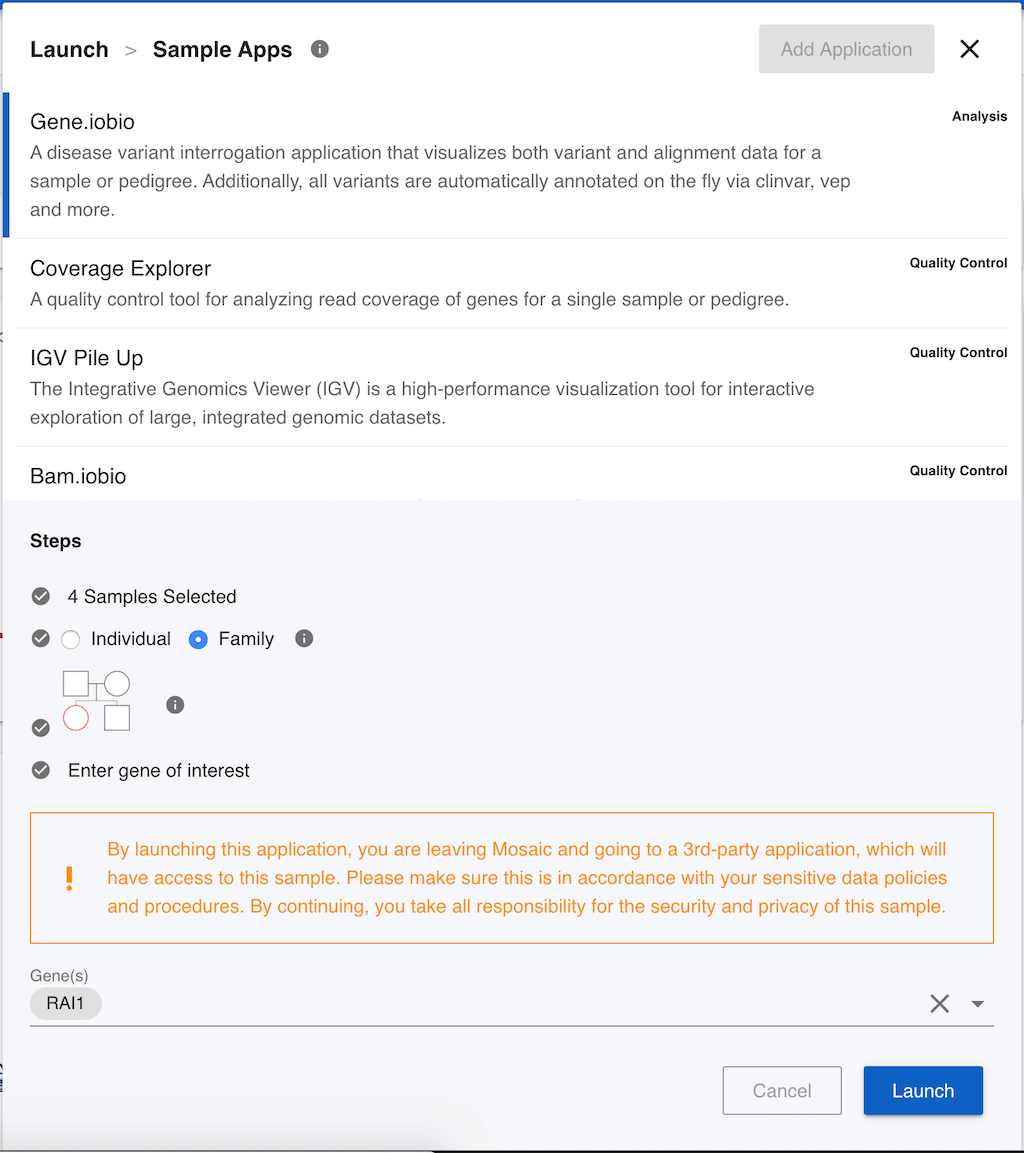
Gene.iobio allows users to interrogate sequencing coverage and genetic variants in families, providing real-time annotation of variants in the genes that you choose. First of all, we have to decide if we want to use gene.iobio for the individual sample, or the whole family. If this sample has been identified as part of a family, Mosaic will default to analyzing the family, so we will leave Family selected. Gene.iobio needs to know who the proband is. This is the sample that we want to interrogate, typically the patient in a clinical analysis, and is always one of the children in gene.iobio. We can click on the pedigree image to change the proband (highlighted in red). As we are on the sample page for NA12878, she is highlighted by default, and we will leave that selected. Finally, we need to select a gene that we want to investigate. For the purposes of demonstration, this data set has had some genetic variants manually inserted, so we can enter the gene name RAI1 (as shown in the image) and click Launch.
Gene.iobio will launch in a new tab and will pull in information on the RAI1 gene for all four samples of the pedigree and show the sequencing coverage and variants for NA12878 - our proband. In real-time, it finds out information about the genetic variants, for example, were they inherited from her parents, and if so, in a recessive or dominant fashion? How common is this variant in the general population (using information from the gnomAD project)? Are any of the variants known to be associated with disease, based on information from the ClinVar project?
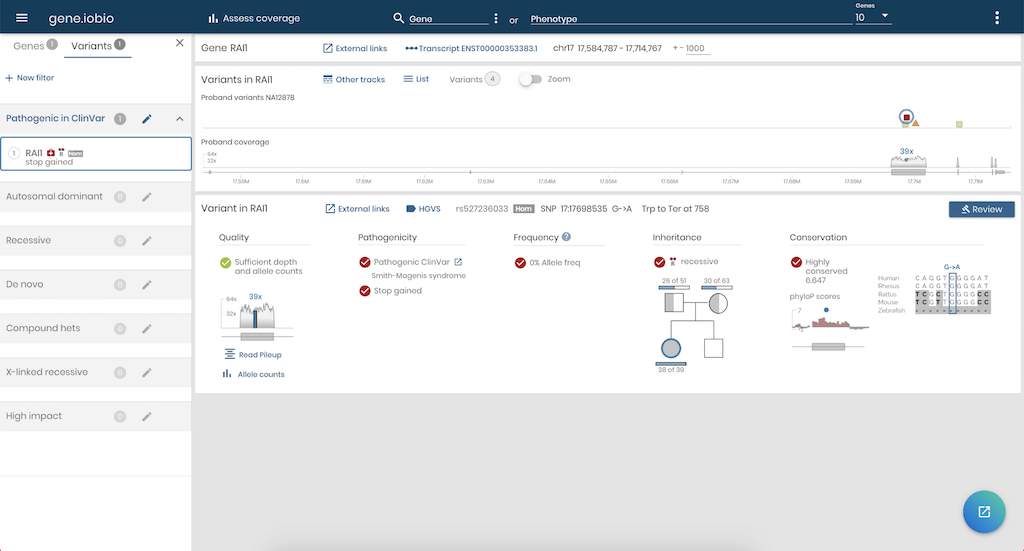
You can choose to look at other genes by typing them into the Gene field in the menu bar, or click on any variant to review information about the variant.
More information on gene.iobio, and other apps are available in the Get Started Guide.